Large RNA including mRNA (mRNA) has emerged as an important new class of therapeutics. Recently, this has been demonstrated by two highly efficacious vaccines based on mRNA sequences encoding for a modified version of the SARS-CoV-2 spike protein. There is currently significant demand for the development of new and improved analytical methods for the characterization of large RNA including mRNA therapeutics.
Researchers from the University of Sheffield have developed an automated, high-throughput workflow for the rapid characterization and direct sequence mapping of large RNA and mRNA therapeutics. Partial RNase digestions using RNase T1 immobilized on magnetic particles were performed in conjunction with high-resolution liquid chromatography-mass spectrometry analysis. Sequence mapping was performed using automated oligoribonucleotide annotation and identifications based on MS/MS spectra. Using this approach, a >80% sequence of coverage of a range of large RNAs and mRNA therapeutics including the SARS-CoV-2 spike protein was obtained in a single analysis. The analytical workflow, including automated sample preparation, can be completed within 90 min. The ability to rapidly identify, characterize, and sequence map large mRNA therapeutics with high sequence coverage provides important information for identity testing, sequence validation, and impurity analysis.
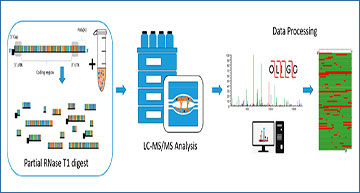
Schematic illustration of mRNA sequence mapping workflow. Partial RNase T1 digests are performed in conjunction with LC–MS/MS analysis for automated oligoribonucleotide annotation and identification prior to sequence mapping.
Vanhinsbergh CJ, Criscuolo A, Sutton JN, Murphy K, Williamson AJK, Cook K, Dickman MJ. (2022) Characterization and Sequence Mapping of Large RNA and mRNA Therapeutics Using Mass Spectrometry. Anal Chem [Epub ahead of print]. [article]