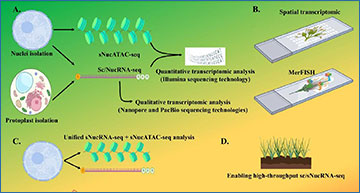
Plant single-cell RNA-seq technology quantifies the abundance of plant transcripts at a single-cell resolution. Deciphering the transcriptomes of each plant cell, their regulation during plant cell development, and their response to environmental stresses will support the functional study of genes, the establishment of precise transcriptional programs, the prediction of more accurate gene regulatory networks, and, in the long term, the design of de novo gene pathways to enhance selected crop traits.
Researchers at the University of Nebraska-Lincoln discuss the opportunities, challenges, and problems, and share tentative solutions associated with the generation and analysis of plant single-cell transcriptomes. The researchers discuss the benefit and limitations of using plant protoplasts vs. nuclei to conduct single-cell RNA-seq experiments on various plant species and organs, the functional annotation of plant cell types based on their transcriptomic profile, the characterization of the dynamic regulation of the plant genes during cell development or in response to environmental stress, the need to characterize and integrate additional layers of –omics datasets to capture new molecular modalities at the single-cell level and reveal their causalities, the deposition and access to single-cell datasets, and the accessibility of this technology to plant scientists.
Cervantes-Pérez SA, Thibivillliers S, Tennant S, Libault M. (2022) Challenges and perspectives in applying single nuclei RNA-seq technology in plant biology. Plant Sci 325:111486. [article]