Single nuclei RNA sequencing (snRNA-seq) has evolved as a powerful tool to study complex human diseases. Single cell resolution enables the study of novel cell types, biological processes, cell trajectories, and cell–cell signaling pathways. snRNA-seq largely relies on the dissociation of intact nuclei from human tissues. However, the study of complex tissues using small core biopsies presents many technical challenges.
Researchers from the University of Maryland School of Medicine have developed an optimized protocol for single nuclei isolation for frozen and RNAlater preserved human kidney biopsies. The described protocol is fast, low cost, and time effective due to the elimination of cell sorting and ultra-centrifugation. Samples can be processed in 90 min or less. This method is effective for obtaining normal nuclei morphology without signs of structural damage. Using snRNA-seq, 16 distinct kidney cell clusters were recovered from normal and peri-transplant acute kidney injury allograft samples, including immune cell clusters. Quality control measurements demonstrated that these optimizations eliminated cellular debris and allowed for a high yield of high-quality nuclei and RNA for library preparation and sequencing. Cellular disassociation did not induce cellular stress responses, which recapitulated transcriptional patterns associated with standardized methods of nuclei isolation. Future applications of this protocol will allow for thorough investigations of small biobank biopsies, identifying cell-specific injury pathways and driving the discovery of novel diagnostics and therapeutic targets.
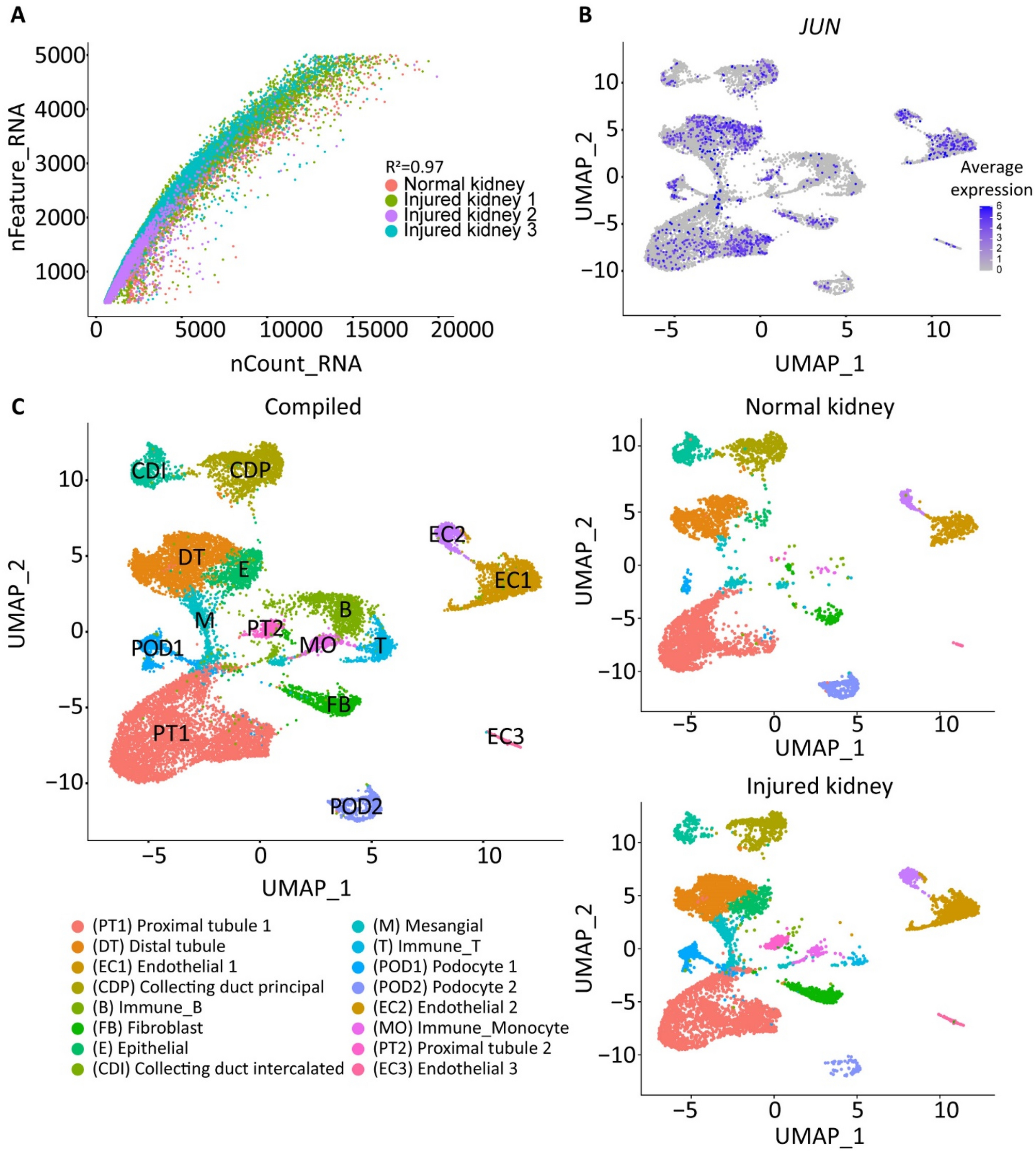
Assessment of single nuclei quality after snRNA-seq
(A) Dot plot representation of the relationship of the number of genes detected (y-axis) against the total number of RNA reads (x-axis). R2 = 0.97. (B) Average expression of JUN stress response pathway across each cell population. (C) (left) UMAP visualization of a total of 13,075 nuclei partitioned into 16 clusters from a total of 5 human kidneys after quality control filtering. Each cluster is labeled by color. (right) Individual UMAP visualization of the normal and injured kidney samples.
Rousselle TV, McDaniels JM, Shetty AC, Bardhi E, Maluf DG, Mas VR. (2022) An optimized protocol for single nuclei isolation from clinical biopsies for RNA-seq. Sci Rep 12(1):9851. [article]