A transcriptome constructed from short-read RNA sequencing (RNA-seq) is an easily attainable proxy catalog of protein-coding genes when genome assembly is unnecessary, expensive or difficult. In the absence of a sequenced genome to guide the reconstruction process, the transcriptome must be assembled de novo using only the information available in the RNA-seq reads. Subsequently, the sequences must be annotated in order to identify sequence-intrinsic and evolutionary features in them (for example, protein-coding regions). Although straightforward at first glance, de novo transcriptome assembly and annotation can quickly prove to be challenging undertakings. In addition to familiarizing themselves with the conceptual and technical intricacies of the tasks at hand and the numerous pre- and post-processing steps involved, those interested must also grapple with an overwhelmingly large choice of tools. The lack of standardized workflows, fast pace of development of new tools and techniques and paucity of authoritative literature have served to exacerbate the difficulty of the task even further. Researchers from the Max Planck Institute for Biophysical Chemistry present a comprehensive overview of de novo transcriptome assembly and annotation. They discuss the procedures involved, including pre- and post-processing steps, and present a compendium of corresponding tools.
Assembly and annotation workflow
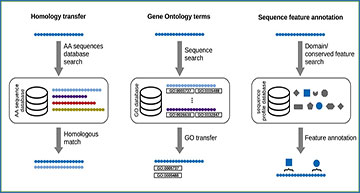
(A) Quality control of the raw reads by filtering for erroneous reads and sequencing artifacts. (B) Sequence assembly including clustering into groups of isoforms and removing redundant sequences (isoforms are transcript variants arising from alternative splicing). (C) Mapping the raw reads to the assembled sequences for either quality control of the assembly or for differential expression analysis. (D) Applying statistical tests for identification of changes in expression levels. (E) Classifying sequences by RNA species and translating into protein sequences before annotation. (F) Annotating sequences on the basis of sequence similarity, identifying sequence features (such as functional domains) and annotating Gene Ontology terms.
Raghavan V, Kraft L, Mesny F, Rigerte L. (2022) A simple guide to de novo transcriptome assembly and annotation. Brief Bioinform 23(2):bbab563. [article]